From Developer to Machine Learning Practitioner in 14 Days
Python is one of the fastest-growing platforms for applied machine learning.
In this mini-course, you will discover how you can get started, build accurate models and confidently complete predictive modeling machine learning projects using Python in 14 days.
This is a big and important post. You might want to bookmark it.
Let’s get started.
![Python Machine Learning Mini-Course]()
Python Machine Learning Mini-Course
Photo by Dave Young, some rights reserved.
Who Is This Mini-Course For?
Before we get started, let’s make sure you are in the right place.
The list below provides some general guidelines as to who this course was designed for.
Don’t panic if you don’t match these points exactly, you might just need to brush up in one area or another to keep up.
- Developers that know how to write a little code. This means that it is not a big deal for you to pick up a new programming language like Python once you know the basic syntax. It does not mean you’re a wizard coder, just that you can follow a basic C-like language with little effort.
- Developers that know a little machine learning. This means you know the basics of machine learning like cross-validation, some algorithms and the bias-variance trade-off. It does not mean that you are a machine learning Ph.D., just that you know the landmarks or know where to look them up.
This mini-course is neither a textbook on Python or a textbook on machine learning.
It will take you from a developer that knows a little machine learning to a developer who can get results using the Python ecosystem, the rising platform for professional machine learning.
Your Guide to Machine Learning with Scikit-Learn
![Python Mini-Course]() Python and scikit-learn are the rising platform among professional data scientists for applied machine learning.
Python and scikit-learn are the rising platform among professional data scientists for applied machine learning.
PDF and Email Course.
FREE 14-Day Mini-Course in
Machine Learning with Python and scikit-learn
Download Your FREE Mini-Course >>
Download your PDF containing all 14 lessons.
Get your daily lesson via email with tips and tricks.
Mini-Course Overview
This mini-course is broken down into 14 lessons.
You could complete one lesson per day (recommended) or complete all of the lessons in one day (hard core!). It really depends on the time you have available and your level of enthusiasm.
Below are 14 lessons that will get you started and productive with machine learning in Python:
- Lesson 1: Download and Install Python and SciPy ecosystem.
- Lesson 2: Get Around In Python, NumPy, Matplotlib and Pandas.
- Lesson 3: Load Data From CSV.
- Lesson 4: Understand Data with Descriptive Statistics.
- Lesson 5: Understand Data with Visualization.
- Lesson 6: Prepare For Modeling by Pre-Processing Data.
- Lesson 7: Algorithm Evaluation With Resampling Methods.
- Lesson 8: Algorithm Evaluation Metrics.
- Lesson 9: Spot-Check Algorithms.
- Lesson 10: Model Comparison and Selection.
- Lesson 11: Improve Accuracy with Algorithm Tuning.
- Lesson 12: Improve Accuracy with Ensemble Predictions.
- Lesson 13: Finalize And Save Your Model.
- Lesson 14: Hello World End-to-End Project.
Each lesson could take you 60 seconds or up to 30 minutes. Take your time and complete the lessons at your own pace. Ask questions and even post results in the comments below.
The lessons expect you to go off and find out how to do things. I will give you hints, but part of the point of each lesson is to force you to learn where to go to look for help on and about the Python platform (hint, I have all of the answers directly on this blog, use the search feature).
I do provide more help in the early lessons because I want you to build up some confidence and inertia.
Hang in there, don’t give up!
Lesson 1: Download and Install Python and SciPy
You cannot get started with machine learning in Python until you have access to the platform.
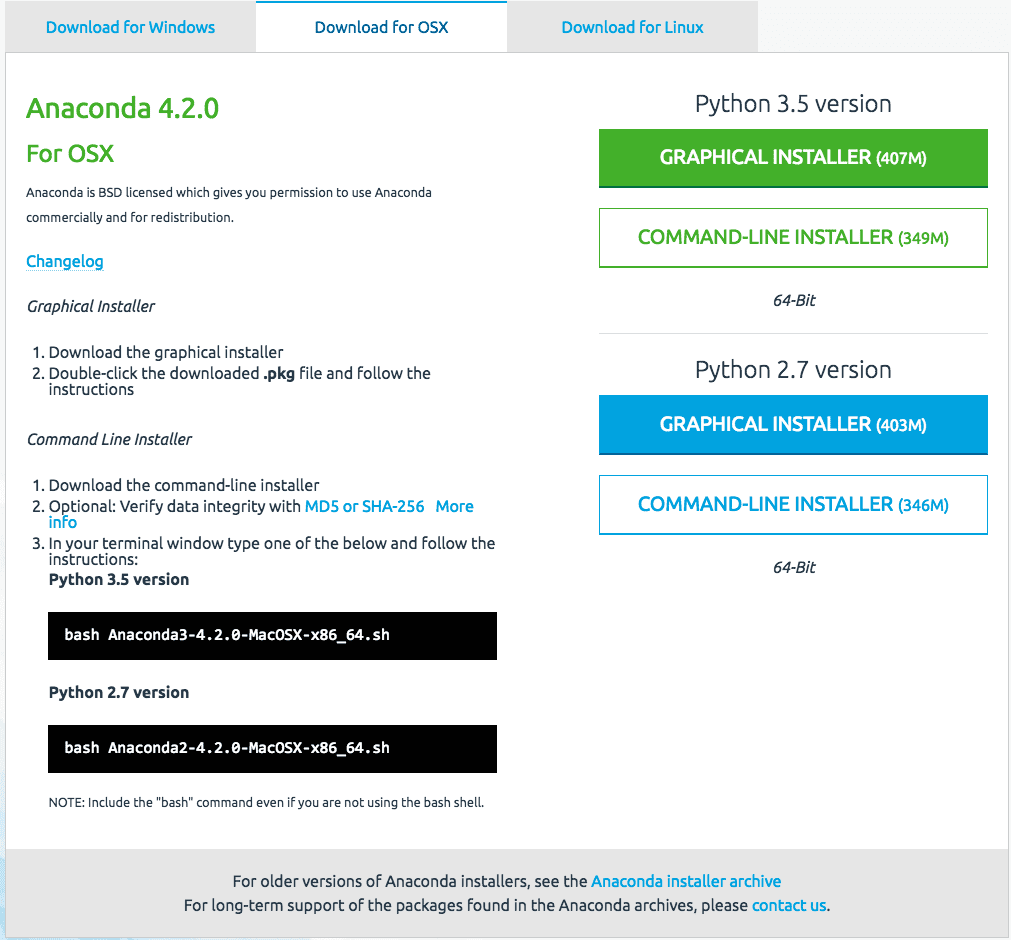
Today’s lesson is easy, you must download and install the Python 2.7 platform on your computer.
Visit the Python homepage and download Python for your operating system (Linux, OS X or Windows). Install Python on your computer. You may need to use a platform specific package manager such as macports on OS X or yum on RedHat Linux.
You also need to install the SciPy platform and the scikit-learn library. I recommend using the same approach that you used to install Python.
You can install everything at once (much easier) with Anaconda. Recommended for beginners.
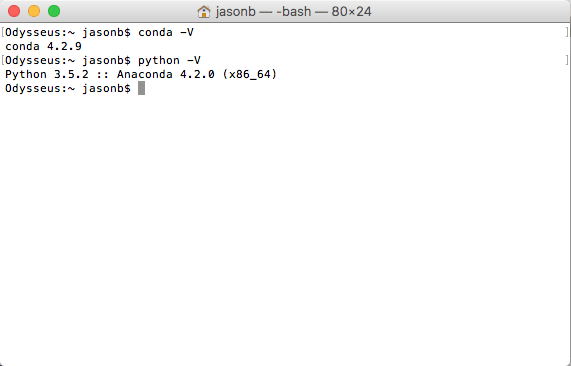
Start Python for the first time by typing “python” at the command line.
Check the versions of everything you are going to need using the code below:
# Python version
import sys
print('Python: {}'.format(sys.version))
# scipy
import scipy
print('scipy: {}'.format(scipy.__version__))
# numpy
import numpy
print('numpy: {}'.format(numpy.__version__))
# matplotlib
import matplotlib
print('matplotlib: {}'.format(matplotlib.__version__))
# pandas
import pandas
print('pandas: {}'.format(pandas.__version__))
# scikit-learn
import sklearn
print('sklearn: {}'.format(sklearn.__version__))If there are any errors, stop.
Now is the time to fix them.
Lesson 2: Get Around In Python, NumPy, Matplotlib and Pandas.
You need to be able to read and write basic Python scripts.
As a developer, you can pick-up new programming languages pretty quickly. Python is case sensitive, uses hash (#) for comments and uses whitespace to indicate code blocks (whitespace matters).
Today’s task is to practice the basic syntax of the Python programming language and important SciPy data structures in the Python interactive environment.
- Practice assignment, working with lists and flow control in Python.
- Practice working with NumPy arrays.
- Practice creating simple plots in Matplotlib.
- Practice working with Pandas Series and DataFrames.
For example, below is a simple example of creating a Pandas DataFrame.
# dataframe
import numpy
import pandas
myarray = numpy.array([[1, 2, 3], [4, 5, 6]])
rownames = ['a', 'b']
colnames = ['one', 'two', 'three']
mydataframe = pandas.DataFrame(myarray, index=rownames, columns=colnames)
print(mydataframe)
Lesson 3: Load Data From CSV
Machine learning algorithms need data. You can load your own data from CSV files but when you are getting started with machine learning in Python you should practice on standard machine learning datasets.
Your task for today’s lesson is to get comfortable loading data into Python and to find and load standard machine learning datasets.
There are many excellent standard machine learning datasets in CSV format that you can download and practice with on the UCI machine learning repository.
- Practice loading CSV files into Python using the CSV.reader() in the standard library.
- Practice loading CSV files using NumPy and the numpy.loadtxt() function.
- Practice loading CSV files using Pandas and the pandas.read_csv() function.
To get you started, below is a snippet that will load the Pima Indians onset of diabetes dataset using Pandas directly from the UCI Machine Learning Repository.
# Load CSV using Pandas from URL
import pandas
url = "https://goo.gl/vhm1eU"
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
data = pandas.read_csv(url, names=names)
print(data.shape)
Well done for making it this far! Hang in there.
Any questions so far? Ask in the comments.
Lesson 4: Understand Data with Descriptive Statistics
Once you have loaded your data into Python you need to be able to understand it.
The better you can understand your data, the better and more accurate the models that you can build. The first step to understanding your data is to use descriptive statistics.
Today your lesson is to learn how to use descriptive statistics to understand your data. I recommend using the helper functions provided on the Pandas DataFrame.
- Understand your data using the head() function to look at the first few rows.
- Review the dimensions of your data with the shape property.
- Look at the data types for each attribute with the dtypes property.
- Review the distribution of your data with the describe() function.
- Calculate pairwise correlation between your variables using the corr() function.
The below example loads the Pima Indians onset of diabetes dataset and summarizes the distribution of each attribute.
# Statistical Summary
import pandas
url = "https://goo.gl/vhm1eU"
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
data = pandas.read_csv(url, names=names)
description = data.describe()
print(description)
Try it out!
Lesson 5: Understand Data with Visualization
Continuing on from yesterdays lesson, you must spend time to better understand your data.
A second way to improve your understanding of your data is by using data visualization techniques (e.g. plotting).
Today, your lesson is to learn how to use plotting in Python to understand attributes alone and their interactions. Again, I recommend using the helper functions provided on the Pandas DataFrame.
- Use the hist() function to create a histogram of each attribute.
- Use the plot(kind=’box’) function to create box-and-whisker plots of each attribute.
- Use the pandas.scatter_matrix() function to create pairwise scatterplots of all attributes.
For example, the snippet below will load the diabetes dataset and create a scatterplot matrix of the dataset.
# Scatter Plot Matrix
import matplotlib.pyplot as plt
import pandas
from pandas.tools.plotting import scatter_matrix
url = "https://goo.gl/vhm1eU"
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
data = pandas.read_csv(url, names=names)
scatter_matrix(data)
plt.show()
![Sample Scatter Plot Matrix]()
Sample Scatter Plot Matrix
Lesson 6: Prepare For Modeling by Pre-Processing Data
Your raw data may not be setup to be in the best shape for modeling.
Sometimes you need to preprocess your data in order to best present the inherent structure of the problem in your data to the modeling algorithms. In today’s lesson, you will use the pre-processing capabilities provided by the scikit-learn.
The scikit-learn library provides two standard idioms for transforming data. Each transform is useful in different circumstances: Fit and Multiple Transform and Combined Fit-And-Transform.
There are many techniques that you can use to prepare your data for modeling. For example, try out some of the following
- Standardize numerical data (e.g. mean of 0 and standard deviation of 1) using the scale and center options.
- Normalize numerical data (e.g. to a range of 0-1) using the range option.
- Explore more advanced feature engineering such as Binarizing.
For example, the snippet below loads the Pima Indians onset of diabetes dataset, calculates the parameters needed to standardize the data, then creates a standardized copy of the input data.
# Standardize data (0 mean, 1 stdev)
from sklearn.preprocessing import StandardScaler
import pandas
import numpy
url = "https://goo.gl/vhm1eU"
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
dataframe = pandas.read_csv(url, names=names)
array = dataframe.values
# separate array into input and output components
X = array[:,0:8]
Y = array[:,8]
scaler = StandardScaler().fit(X)
rescaledX = scaler.transform(X)
# summarize transformed data
numpy.set_printoptions(precision=3)
print(rescaledX[0:5,:])
Lesson 7: Algorithm Evaluation With Resampling Methods
The dataset used to train a machine learning algorithm is called a training dataset. The dataset used to train an algorithm cannot be used to give you reliable estimates of the accuracy of the model on new data. This is a big problem because the whole idea of creating the model is to make predictions on new data.
You can use statistical methods called resampling methods to split your training dataset up into subsets, some are used to train the model and others are held back and used to estimate the accuracy of the model on unseen data.
Your goal with today’s lesson is to practice using the different resampling methods available in scikit-learn, for example:
- Split a dataset into training and test sets.
- Estimate the accuracy of an algorithm using k-fold cross validation.
- Estimate the accuracy of an algorithm using leave one out cross validation.
The snippet below uses scikit-learn to estimate the accuracy of the Logistic Regression algorithm on the Pima Indians onset of diabetes dataset using 10-fold cross validation.
# Evaluate using Cross Validation
import pandas
from sklearn import cross_validation
from sklearn.linear_model import LogisticRegression
url = "https://goo.gl/vhm1eU"
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
dataframe = pandas.read_csv(url, names=names)
array = dataframe.values
X = array[:,0:8]
Y = array[:,8]
num_folds = 10
num_instances = len(X)
seed = 7
kfold = cross_validation.KFold(n=num_instances, n_folds=num_folds, random_state=seed)
model = LogisticRegression()
results = cross_validation.cross_val_score(model, X, Y, cv=kfold)
print("Accuracy: %.3f%% (%.3f%%)") % (results.mean()*100.0, results.std()*100.0)What accuracy did you get? Let me know in the comments.
Did you realize that this is the halfway point? Well done!
Lesson 8: Algorithm Evaluation Metrics
There are many different metrics that you can use to evaluate the skill of a machine learning algorithm on a dataset.
You can specify the metric used for your test harness in scikit-learn via the cross_validation.cross_val_score() function and defaults can be used for regression and classification problems. Your goal with today’s lesson is to practice using the different algorithm performance metrics available in the scikit-learn package.
- Practice using the Accuracy and LogLoss metrics on a classification problem.
- Practice generating a confusion matrix and a classification report.
- Practice using RMSE and RSquared metrics on a regression problem.
The snippet below demonstrates calculating the LogLoss metric on the Pima Indians onset of diabetes dataset.
# Cross Validation Classification LogLoss
import pandas
from sklearn import cross_validation
from sklearn.linear_model import LogisticRegression
url = "https://goo.gl/vhm1eU"
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
dataframe = pandas.read_csv(url, names=names)
array = dataframe.values
X = array[:,0:8]
Y = array[:,8]
num_folds = 10
num_instances = len(X)
seed = 7
kfold = cross_validation.KFold(n=num_instances, n_folds=num_folds, random_state=seed)
model = LogisticRegression()
scoring = 'log_loss'
results = cross_validation.cross_val_score(model, X, Y, cv=kfold, scoring=scoring)
print("Logloss: %.3f (%.3f)") % (results.mean(), results.std())What log loss did you get? Let me know in the comments.
Lesson 9: Spot-Check Algorithms
You cannot possibly know which algorithm will perform best on your data beforehand.
You have to discover it using a process of trial and error. I call this spot-checking algorithms. The scikit-learn library provides an interface to many machine learning algorithms and tools to compare the estimated accuracy of those algorithms.
In this lesson, you must practice spot checking different machine learning algorithms.
- Spot check linear algorithms on a dataset (e.g. linear regression, logistic regression and linear discriminate analysis).
- Spot check some non-linear algorithms on a dataset (e.g. KNN, SVM and CART).
- Spot-check some sophisticated ensemble algorithms on a dataset (e.g. random forest and stochastic gradient boosting).
For example, the snippet below spot-checks the K-Nearest Neighbors algorithm on the Boston House Price dataset.
# KNN Regression
import pandas
from sklearn import cross_validation
from sklearn.neighbors import KNeighborsRegressor
url = "https://goo.gl/sXleFv"
names = ['CRIM', 'ZN', 'INDUS', 'CHAS', 'NOX', 'RM', 'AGE', 'DIS', 'RAD', 'TAX', 'PTRATIO', 'B', 'LSTAT', 'MEDV']
dataframe = pandas.read_csv(url, delim_whitespace=True, names=names)
array = dataframe.values
X = array[:,0:13]
Y = array[:,13]
num_folds = 10
num_instances = len(X)
seed = 7
kfold = cross_validation.KFold(n=num_instances, n_folds=num_folds, random_state=seed)
model = KNeighborsRegressor()
scoring = 'mean_squared_error'
results = cross_validation.cross_val_score(model, X, Y, cv=kfold, scoring=scoring)
print(results.mean())
What mean squared error did you get? Let me know in the comments.
Lesson 10: Model Comparison and Selection
Now that you know how to spot check machine learning algorithms on your dataset, you need to know how to compare the estimated performance of different algorithms and select the best model.
In today’s lesson, you will practice comparing the accuracy of machine learning algorithms in Python with scikit-learn.
- Compare linear algorithms to each other on a dataset.
- Compare nonlinear algorithms to each other on a dataset.
- Compare different configurations of the same algorithm to each other.
- Create plots of the results comparing algorithms.
The example below compares Logistic Regression and Linear Discriminant Analysis to each other on the Pima Indians onset of diabetes dataset.
# Compare Algorithms
import pandas
from sklearn import cross_validation
from sklearn.linear_model import LogisticRegression
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
# load dataset
url = "https://goo.gl/vhm1eU"
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
dataframe = pandas.read_csv(url, names=names)
array = dataframe.values
X = array[:,0:8]
Y = array[:,8]
# prepare configuration for cross validation test harness
num_folds = 10
num_instances = len(X)
seed = 7
# prepare models
models = []
models.append(('LR', LogisticRegression()))
models.append(('LDA', LinearDiscriminantAnalysis()))
# evaluate each model in turn
results = []
names = []
scoring = 'accuracy'
for name, model in models:
kfold = cross_validation.KFold(n=num_instances, n_folds=num_folds, random_state=seed)
cv_results = cross_validation.cross_val_score(model, X, Y, cv=kfold, scoring=scoring)
results.append(cv_results)
names.append(name)
msg = "%s: %f (%f)" % (name, cv_results.mean(), cv_results.std())
print(msg)Which algorithm got better results? Can you do better? Let me know in the comments.
Lesson 11: Improve Accuracy with Algorithm Tuning
Once you have found one or two algorithms that perform well on your dataset, you may want to improve the performance of those models.
One way to increase the performance of an algorithm is to tune its parameters to your specific dataset.
The scikit-learn library provides two ways to search for combinations of parameters for a machine learning algorithm. Your goal in today’s lesson is to practice each.
- Tune the parameters of an algorithm using a grid search that you specify.
- Tune the parameters of an algorithm using a random search.
The snippet below uses is an example of using a grid search for the Ridge Regression algorithm on the Pima Indians onset of diabetes dataset.
# Grid Search for Algorithm Tuning
import pandas
import numpy
from sklearn import cross_validation
from sklearn.linear_model import Ridge
from sklearn.grid_search import GridSearchCV
url = "https://goo.gl/vhm1eU"
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
dataframe = pandas.read_csv(url, names=names)
array = dataframe.values
X = array[:,0:8]
Y = array[:,8]
alphas = numpy.array([1,0.1,0.01,0.001,0.0001,0])
param_grid = dict(alpha=alphas)
model = Ridge()
grid = GridSearchCV(estimator=model, param_grid=param_grid)
grid.fit(X, Y)
print(grid.best_score_)
print(grid.best_estimator_.alpha)
Which parameters achieved the best results? Can you do better? Let me know in the comments.
Lesson 12: Improve Accuracy with Ensemble Predictions
Another way that you can improve the performance of your models is to combine the predictions from multiple models.
Some models provide this capability built-in such as random forest for bagging and stochastic gradient boosting for boosting. Another type of ensembling called voting can be used to combine the predictions from multiple different models together.
In today’s lesson, you will practice using ensemble methods.
- Practice bagging ensembles with the random forest and extra trees algorithms.
- Practice boosting ensembles with the gradient boosting machine and AdaBoost algorithms.
- Practice voting ensembles using by combining the predictions from multiple models together.
The snippet below demonstrates how you can use the Random Forest algorithm (a bagged ensemble of decision trees) on the Pima Indians onset of diabetes dataset.
# Random Forest Classification
import pandas
from sklearn import cross_validation
from sklearn.ensemble import RandomForestClassifier
url = "https://goo.gl/vhm1eU"
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
dataframe = pandas.read_csv(url, names=names)
array = dataframe.values
X = array[:,0:8]
Y = array[:,8]
num_folds = 10
num_instances = len(X)
seed = 7
num_trees = 100
max_features = 3
kfold = cross_validation.KFold(n=num_instances, n_folds=num_folds, random_state=seed)
model = RandomForestClassifier(n_estimators=num_trees, max_features=max_features)
results = cross_validation.cross_val_score(model, X, Y, cv=kfold)
print(results.mean())
Can you devise a better ensemble? Let me know in the comments.
Lesson 13: Finalize And Save Your Model
Once you have found a well-performing model on your machine learning problem, you need to finalize it.
In today’s lesson, you will practice the tasks related to finalizing your model.
Practice making predictions with your model on new data (data unseen during training and testing).
Practice saving trained models to file and loading them up again.
For example, the snippet below shows how you can create a Logistic Regression model, save it to file, then load it later and make predictions on unseen data.
# Save Model Using Pickle
import pandas
from sklearn import cross_validation
from sklearn.linear_model import LogisticRegression
import pickle
url = "https://goo.gl/vhm1eU"
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
dataframe = pandas.read_csv(url, names=names)
array = dataframe.values
X = array[:,0:8]
Y = array[:,8]
test_size = 0.33
seed = 7
X_train, X_test, Y_train, Y_test = cross_validation.train_test_split(X, Y, test_size=test_size, random_state=seed)
# Fit the model on 33%
model = LogisticRegression()
model.fit(X_train, Y_train)
# save the model to disk
filename = 'finalized_model.sav'
pickle.dump(model, open(filename, 'wb'))
# some time later...
# load the model from disk
loaded_model = pickle.load(open(filename, 'rb'))
result = loaded_model.score(X_test, Y_test)
print(result)
Lesson 14: Hello World End-to-End Project
You now know how to complete each task of a predictive modeling machine learning problem.
In today’s lesson, you need to practice putting the pieces together and working through a standard machine learning dataset end-to-end.
Work through the iris dataset end-to-end (the hello world of machine learning)
This includes the steps:
- Understanding your data using descriptive statistics and visualization.
- Preprocessing the data to best expose the structure of the problem.
- Spot-checking a number of algorithms using your own test harness.
- Improving results using algorithm parameter tuning.
- Improving results using ensemble methods.
- Finalize the model ready for future use.
Take it slowly and record your results along the way.
What model did you use? What results did you get? Let me know in the comments.
The End!
(Look How Far You Have Come)
You made it. Well done!
Take a moment and look back at how far you have come.
- You started off with an interest in machine learning and a strong desire to be able to practice and apply machine learning using Python.
- You downloaded, installed and started Python, perhaps for the first time and started to get familiar with the syntax of the language.
- Slowly and steadily over the course of a number of lessons you learned how the standard tasks of a predictive modeling machine learning project map onto the Python platform.
- Building upon the recipes for common machine learning tasks you worked through your first machine learning problems end-to-end using Python.
- Using a standard template, the recipes and experience you have gathered you are now capable of working through new and different predictive modeling machine learning problems on your own.
Don’t make light of this, you have come a long way in a short amount of time.
This is just the beginning of your machine learning journey with Python. Keep practicing and developing your skills.
Summary
How Did You Go With The Mini-Course?
Did you enjoy this mini-course?
Do you have any questions? Were there any sticking points?
Let me know. Leave a comment below.
Frustrated With Python Machine Learning?
Develop Your Own Models and Predictions in Minutes
...with just a few lines of scikit-learn code
Discover how in my new Ebook: Machine Learning Mastery With Python
It covers self-study tutorials and end-to-end projects on topics like:
Loading data, visualization, modeling, algorithm tuning, and much more...
Finally Bring Machine Learning To
Your Own Projects
Skip the Academics. Just Results.
Click to learn more.
The post Python Machine Learning Mini-Course appeared first on Machine Learning Mastery.

 Python and scikit-learn are the rising platform among professional data scientists for applied machine learning.
Python and scikit-learn are the rising platform among professional data scientists for applied machine learning. Discover how to confidently step-through machine learning projects end-to-end in Python with scikit-learn in the new Ebook:
Discover how to confidently step-through machine learning projects end-to-end in Python with scikit-learn in the new Ebook: